Working with Jupyter and PySpark
PySpark is a Python-based language for writing distributed programs that can be computed on a Spark cluster (Dataproc Hadoop). PySpark is an alternative to the BigQuery engine for querying and manipulating big data.
Being able to run each step independently and interactively means you can test and refine each step of your code before moving on to the next step.
PySpark provides an advanced data structure (dataframe), which supports a wide range of useful operations on large collections of data. Users can interactively run their code on the remote Dataproc cluster through Jupyter Notebooks by using Sparkmagic.
Note
JupyterLab and Jupyter Notebook can be opened from the Analytics Environment virtual machine desktop.
Jupyter and PySpark can be used for the following activities:
Exploring your data using Python or PySpark libraries
Running short-duration Python or PySpark code
Preparing/testing your code with a subset of data before running it with a huge amount of data
The following diagram illustrates the relationships between Jupyter, PySpark, and BigQuery:
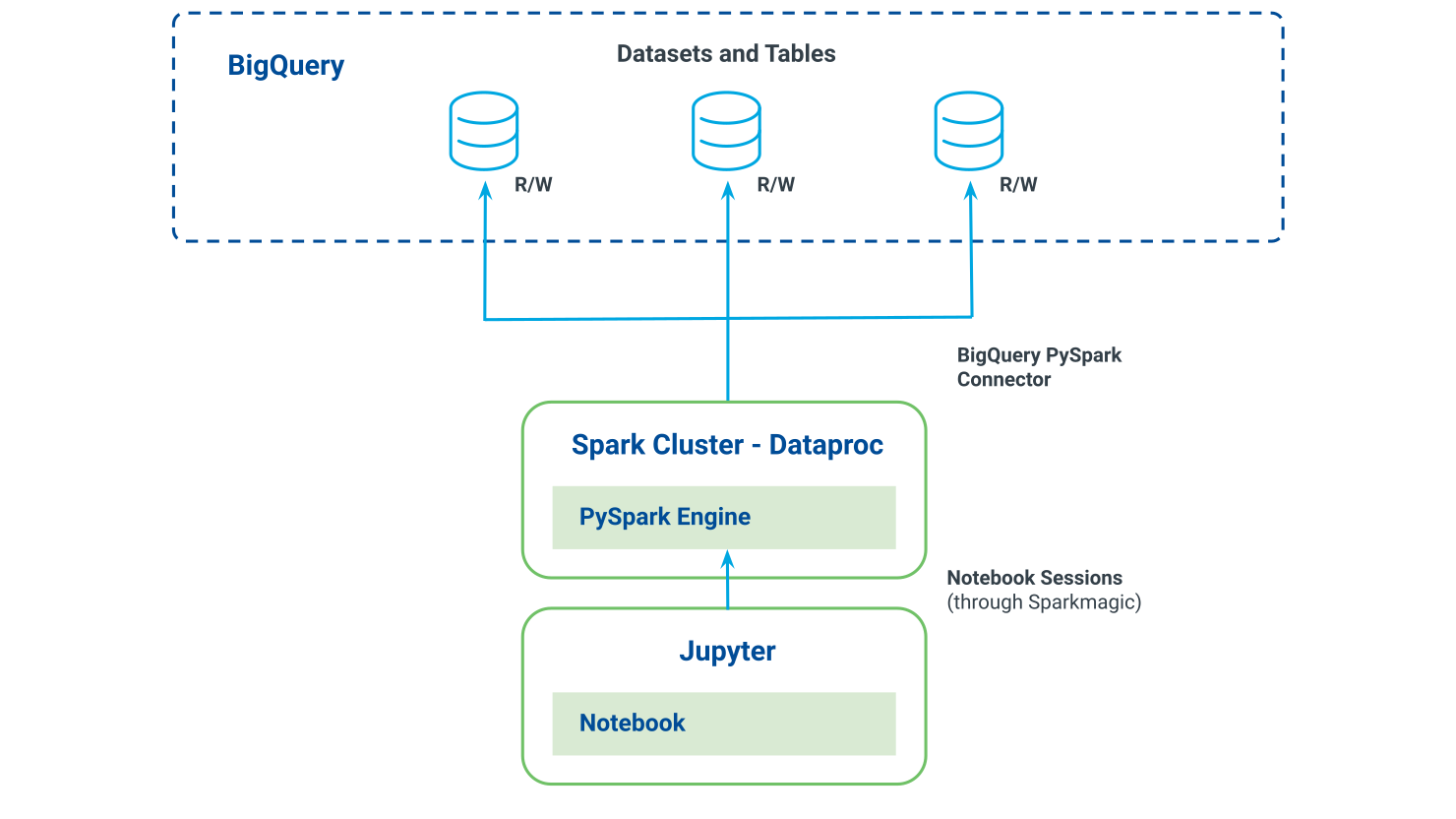
Connect to the Dataproc cluster and start a Spark session: Use the Sparkmagic command on the first lines of your notebook to set a session from your Jupyter Notebook to the remote Spark cluster. See "Create a PySpark Session."
Read data from BigQuery to a PySpark dataframe: Use BigQuery connector for loading data from BigQuery tables into the Spark cluster (on memory). This process is distributed through all the cluster nodes.
Manipulate and transform your PySpark dataframe: To apply your manipulation on your dataframe, you can use libraries such as PySpark SQL. A transformation is only executed when you read or write a dataframe.
Write the aggregated data to BigQuery: Write back to BigQuery the result of the transformations on your original dataframe or a new dataframe created from the last one.
Reading data from BigQuery directly as a PySpark dataframe is a more efficient and recommended option for building a dataframe from BigQuery (for datasets smaller than 128 MB):
Eliminates the need to create a resilient distributed dataset (RDD) and convert it to a dataframe
Uses BigQuery metadata directly
Cheaper: less processing time and resources
Highly recommended by Google
Steps:
Build the query within a variable. Include any filters and conditions at this stage and load the result in a PySpark dataframe.
QUERY = """ SELECT * FROM ` project_id.dataset_name.table_name ` LIMIT 200 """ bq = bigquery.Client() query_job = bq.query(QUERY) query_job.result() df = spark.read.format('bigquery') \ .option('dataset', query_job.destination.dataset_id)\ .option('table', query_job.destination.table_id) \ .load()Load all of the tables into a PySpark dataframe and manipulate data later with PySpark.sql.
df = spark.read.format('bigquery') \ .option('dataset', "dataset_name")\ .option('table', "table_name") \ .load()Note
PySpark doesn't read data at this stage from BigQuery.
Write the result of your query directly to a table in BigQuery.
QUERY = """ SELECT * FROM ` project_id.dataset_name.table_name ` LIMIT 200 """ bq = bigquery.Client() job_config = bigquery.QueryJobConfig() job_config.destination = bq.dataset("dataset_work").table("tmp_table") query_job = bq.query(QUERY, job_config = job_config) query_job.result() df = spark.read.format('bigquery') \ .option('dataset', query_job.destination.dataset_id )\ .option('table', query_job.destination.table_id ) \ .load()Option #1: Write PySpark dataframe directly to a BigQuery table
df.write.format('bigquery') \ .option('dataset', "dataset_output")\ .option('table', "table_name") \ .save()Option #2: Use a temporary bucket before writing to BigQuery
bucket = spark.sparkContext._jsc.hadoopConfiguration().get('fs.gs.system.bucket') spark.conf.set("temporaryGcsBucket", bucket) df.write.format('bigquery') \ .option('dataset', "dataset_output" )\ .option('table', "table_name" ) \ .save()Tip
A best practice is to use a temporary bucket while loading to BigQuery.
Creating a dataframe where query results are larger than 128 MB:
Query results larger than 128 MB result in errors.
Eliminate errors by configuring a temp table to hold the dataframe data for the duration of the job.
Do not use the "_ai" dataset for the temp table. Use "_work."
Tip
Remember to drop the temp table after all execution and the final result is committed.
Reading data from BigQuery as RDD and converting RDD to PySpark dataframe is an alternative way for building PySpark dataframe from BigQuery (not recommended). Reading the BigQuery table this way would only allow reading the entire table without limit or filter, independent of how big the table is. Further filtering could be done on the resulting dataframe.
Caution
JSON-RDD is loaded into cluster memory with metadata for each record. This process consumes a lot of cluster memory
Tip
A best practice is to use a temporary bucket while loading to BigQuery.
Option #1: Use a temporary bucket while loading to BigQuery
bucket = spark.sparkContext._jsc.hadoopConfiguration().get('fs.gs.system.bucket')
spark.conf.set('temporaryGcsBucket', bucket)
df.write.format('bigquery') \
.option('dataset', "dataset_output")\
.option('table', "table_name") \
.save()Option #2: Write PySpark dataframe directly to BigQuery
df.write.format('bigquery') \
.option('dataset', "dataset_output")\
.option('table', "table_name") \
.save()When working with Jupyter and PySpark, consider the following guidelines.
You can use BigQuery connector to load data from BigQuery tables into a Spark cluster for distributed processing using PySpark. However, BigQuery connector doesn't support reading less than 10 MB. If you want to read less than 10 MB of data, use the following query to read the data directly into a Python dataframe:
QUERY = """ SELECT * FROM dataset.table LIMIT 20 """ df = client.query(QUERY).to_dataframe()
Use PySpark dataframes instead of RDD.
Avoid showing large data in Jupyter. During data manipulation, avoid commands such as “show()”, thus reading the data from BigQuery.
Avoid converting large datasets to Python components.
Before converting a PySpark dataframe to Pandas, verify that it is aggregated. Converting a PySpark dataframe to Pandas will result in processing running on a single node.
To avoid negatively impacting performance and computing capacity, first perform all data manipulation with PySpark or BigQuery before using any needed Python libraries (such as Pandas, Scipy, or Scikit-learn).